New genomic tools
25 05 2022
Category: IM Seminar
Next Monday on 30st of May at 2 pm Mikołaj Dziurzyński from the Department of Environmental Microbiology and Biotechnology will present a seminar entitled „ Development of novel tools for genomic and metagenomic analyses of metal resistant bacteria”. You are welcomed to join!
Abstract:
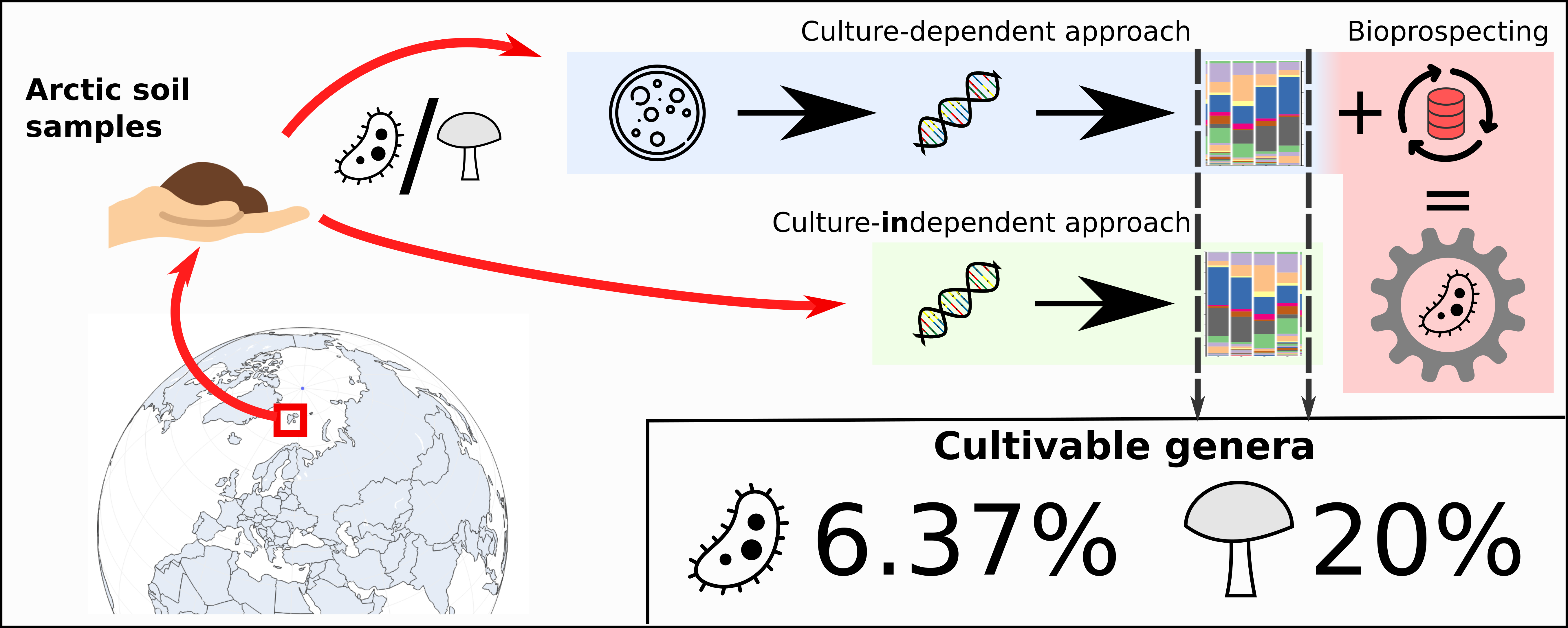
In this work, novel bioinformatics tools, dedicated to analysis of bacterial metal metabolism and resistance, were developed. Each tool has been validated or already successfully used in research. Arctic microbiomes, due to their unexplored uniqueness and high biotechnological potential, were used as an environment of choice for the development, testing and validation of these tools.
The research provided a detailed insight into the biodiversity of Arctic soils, and a first time estimate of uncultivable microbiota present in this environment. Our results showed that standard culturing methods introduce a heavy bias into investigated samples biodiversity and are able to recover only a small portion of bacterial and fungal genera observed using culture-independent methods. Therefore, in order to explore the biotechnological potential of Arctic soils, special, targeted protocols should be used. One such novel protocol was proposed and tested in this study. The protocol relied on relatively inexpensive biodiversity analysis and reuse of genomic data from public repositories, and was designed to increase the chance of isolating multi-metal-resistant bacteria.
A new tool, MAISEN, was developed to increase the efficiency of semi-automatic bacterial DNA annotation. MAISEN allows its users to annotate their sequences of interest using a free and intuitive interface and is available through a web-browser. It allows for annotation of newly sequenced data, as well as for reannotation of already annotated sequences. The main reason behind MAISEN development was to decrease the time needed for accurate manual DNA sequence annotation by providing its users with all-in-one tool that serves precomputed sequence alignments from different databases, so the user can focus on the annotation, and not waste time to running genes, one by one, through different tools and databases.
Finally, the LCPDb-MET database, containing rankings of PCR primers designed to detect genes involved in metal metabolism and resistance was developed. PCR primer benchmark analysis showed that an overwhelming number of analyzed primer pairs was unsuitable for use in an environmental setup. As PCR primer pairs testing required an accurate reference database, and so a novel database (MetGeneDb), proposing an additional level of differentiation for sequences of genes involved in metal metabolism and resistance, was constructed.
