Multiple parS centromere-like sites in repABC replicons
17 05 2022
Category: IM Seminar
You are invited to the institute seminar: next Monday, 23trd of May at 2 pm MSc mgr Elvira Chapkauskaitse from the Department of Bacterial Genetics will present a seminar on „: Multiple parS centromere-like sites in repABC replicons: plasmid pAMI4 of Paracoccus aminophilus JCM 7686 as a case study”.
Abstract:
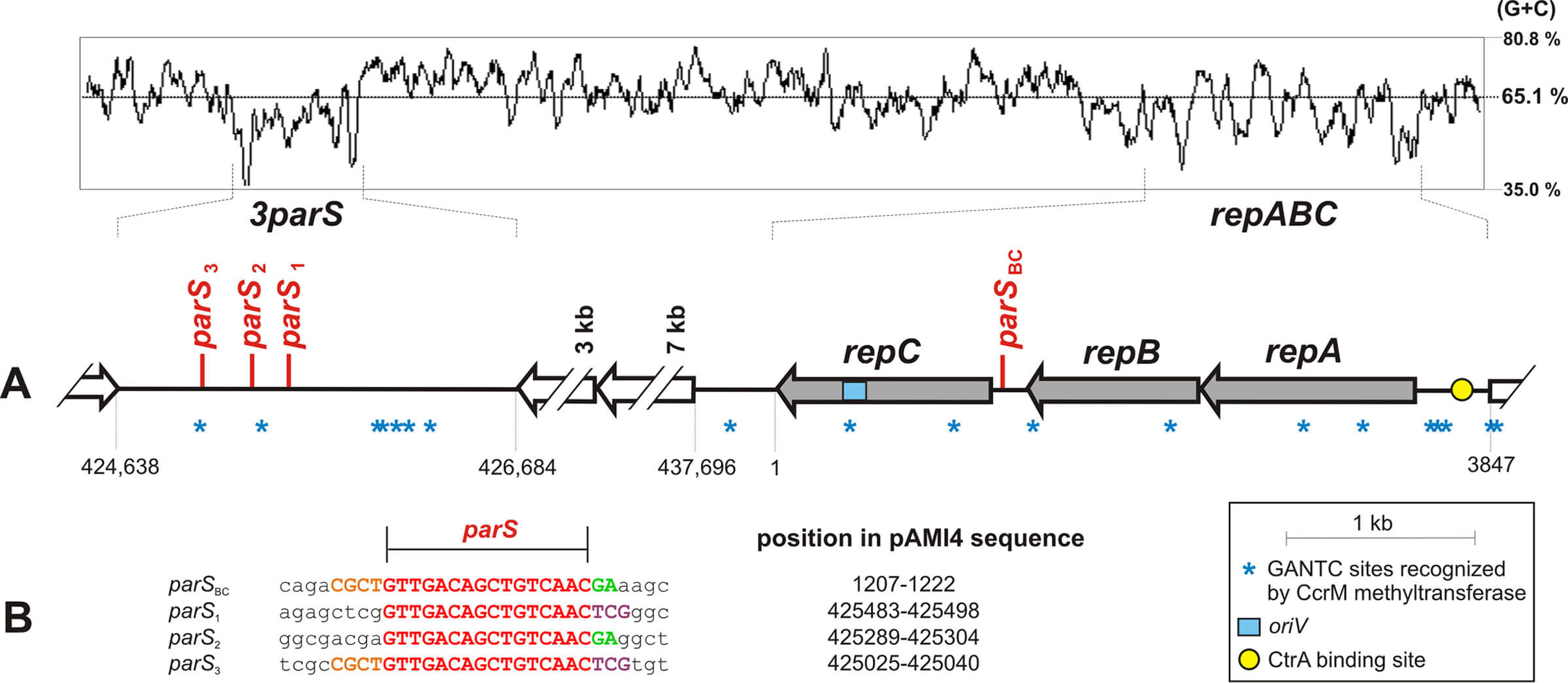
Partitioning systems (par) enable the stable maintenance of bacterial low-copy-number replicons. These loci consist of two genes encoding partition proteins A and B, and a parS centromere-like sequence. The par system is often located in close vicinity to the replication system. An extreme example of this co-localization are alphaproteobacterial repABC replicons, where the partition (repAB) and replication (repC) genes form a single operon, with parS sequences usually positioned in close proximity to these genes. Genomic analysis of Paracoccus aminophilus plasmid pAMI4 (438 kb) revealed that this replicon besides the repABC operon (with parS located between repB and repC) contains a 2-kb-long non-coding locus, carrying three additional parS repeats. The main objective of our study was to examine whether the 3parS locus is involved in pAMI4 maintenance.
We demonstrated that 3parS is bound by partition protein RepB in vitro and is essential for proper pAMI4 partitioning in vivo. In search of similar loci, we conducted a comparative analysis of parS distribution in other repABC replicons. This revealed different patterns of parS localization in Rhodobacterales and Rhizobiales. Many of these additional parS repeats are located inside protein coding sequences, with an interesting example being the cas genes of CRIPSR-Cas systems.
To sum up, we identified a novel locus required for proper maintenance of a repABC plasmid. Our findings raise questions concerning the biological reasons for differential parS distribution, which may reflect variations in repABC operon regulation as well as different replication and partition modes of replicons belonging to the repABC family.
