Domestication of plasmids in Cronobacter
11 03 2025
Category: IM Seminar
On Monday 17th of March at 2 pm in room 102B, MSc Zofia Owczarzak from the Department of Bacterial Genetics will present a seminar entitled “Domestication of mobile DNA – the case study of the virulence plasmids of bacteria of the genus Cronobacter”.
Abstract:
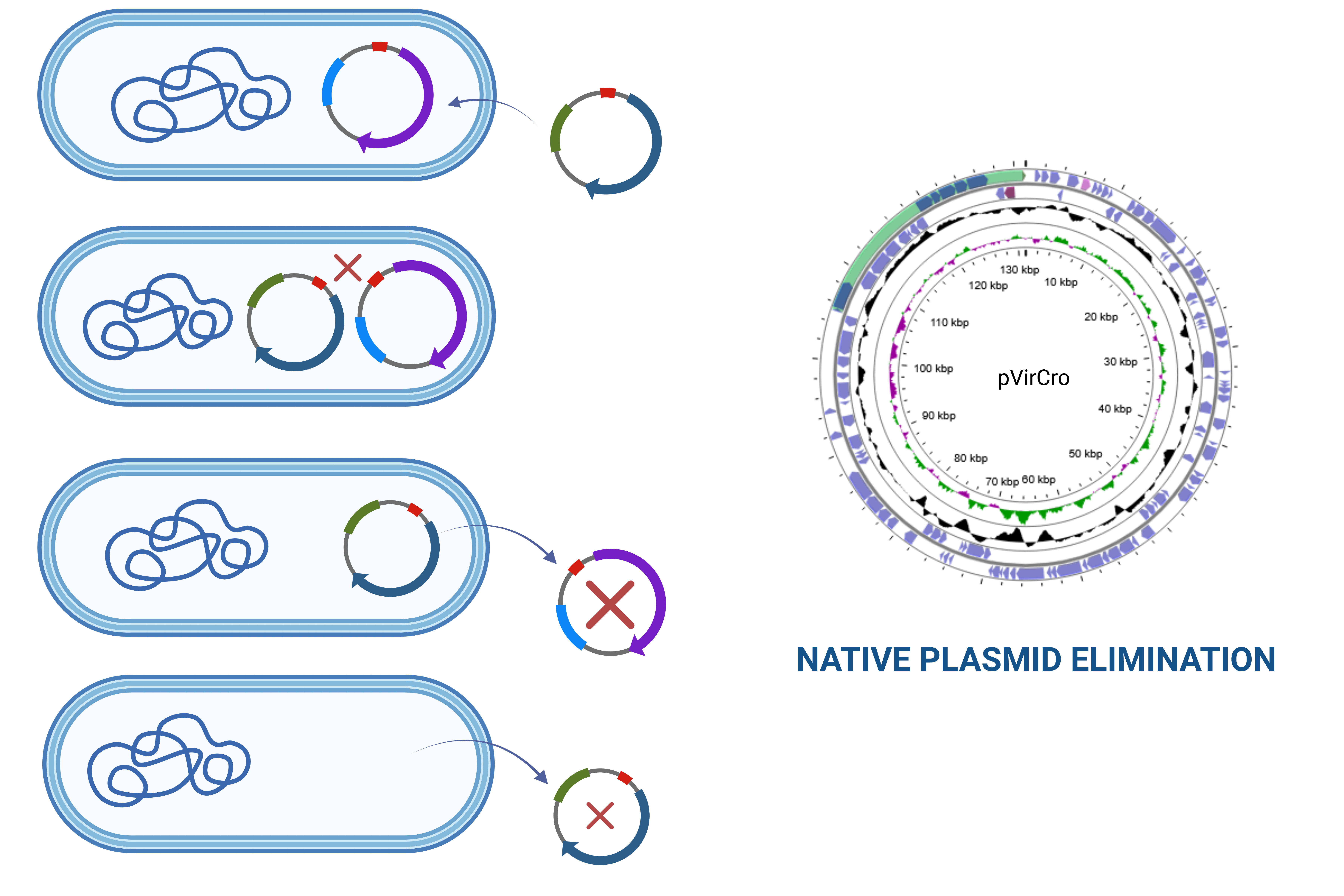
Virulence plasmids of pESA3 family (pVirCro) are present across all Cronobacter species. Studies show their role in adaptability and pathogenicity of this genus since pVirCro encode crucial virulence factors responsible for i.a. biofilm forming abilities and stress response mechanisms. However these replicons have characteristics unusual for plasmids. Their highly conserved backbone and features suggesting coupling of their replication with chromosomes make them similar to chromids. Another notable feature of pVirCro is their genus specificity. These replicons are absent in other taxonomic groups, prevalent in 97% of Cronobacter spp. strains and form a distinct “Cronobacter cluster on the phylogenic tree of RepFIB plasmids. This suggests that Cronobacter genus “domesticated” pVirCro replicons during the course of its evolution.
The study aims to test hypotheses regarding the regulatory coupling of plasmid and chromosomal DNA replication and the underestimated role of these plasmids in host fitness and virulence. Key objectives include elucidating the role of plasmid loci in bacterial adaptation and pathogenicity, understanding the evolutionary significance of plasmid-chromosome interactions, and identifying novel loci linked to virulence. The findings are expected to deepen the understanding of mobile DNA evolution, bacterial multireplicon genomes, and the clinical implications of Cronobacter spp. as opportunistic pathogens. This research holds potential for advancing medical microbiology and addressing challenges related to bacterial infections.
