Holistic insight into the Alphaproteobacteria repABC replicons
07 10 2024
Category: IM Seminar
We would like to invite you to the institute seminar: on Monday 14th of October at 14.15 in 102B room, MSc Elvira Krakowska from the Department of Bacterial Genetics will present a seminar entitled “One Palindrome to Rule Them All: Anchoring distant parS sequences in the Alphaproteobacteria repABC maintenance system”.
Abstract:
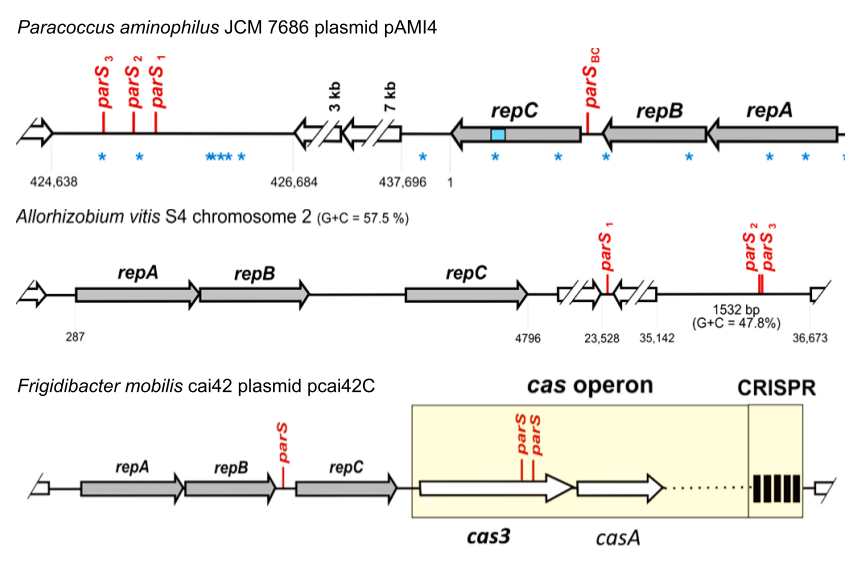
The genomes of many Alphaproteobacteria have multireplicon structure. Besides the chromosome, they often contain large plasmids and megaplasmids as well as secondary chromosomes (chromids). The maintenance of these replicons is very often dependent on repABC operons, encoding proteins providing stabilization (RepA and RepB) and replication (RepC) functions. The repA and repB genes, together with a centromere-like sequence parS, constitute a PAR system, responsible for active partitioning of newly replicated replicon copies during cell division. It is believed that repABC operons contain the complete genetic information necessary to ensure stable maintenance of large replicons. However, we found that numerous replicons contain additional distantly placed parS sites, many of which are located within gene coding sequences. Our preliminary analyses strongly suggest that such additional loci are necessary for proper replicon partitioning. My PhD project aims to analyze biological role the additional parSs in three different types of repABC replicons – plasmid pcai42C of Frigidibacter mobilis, megaplasmid pAMI4 of Paracoccus aminophilus and chromid of Allorhizobium vitis. The project addresses the important questions regarding (1) the role of parS sites in the regulation of expression of cas genes, (2) the relationship between genomic localization of parS sites and the directionality of DNA replication, and (3) the role of distant parSs in segregation of chromid molecules. To conclude, this is the first holistic approach to the stable maintenance of repABC replicons, not limited to repABC operons alone.
